Introduction
Viruses are important component of the human gut microbiota and have a key role in maintaining the well-being of the gut function. However, the role of viruses are neglected in the extensively microbiome-based studies which mainly focused on bacteria. Thus, the role of viruses in the microbial community remains incompletely understood, especially in the context of diseases.
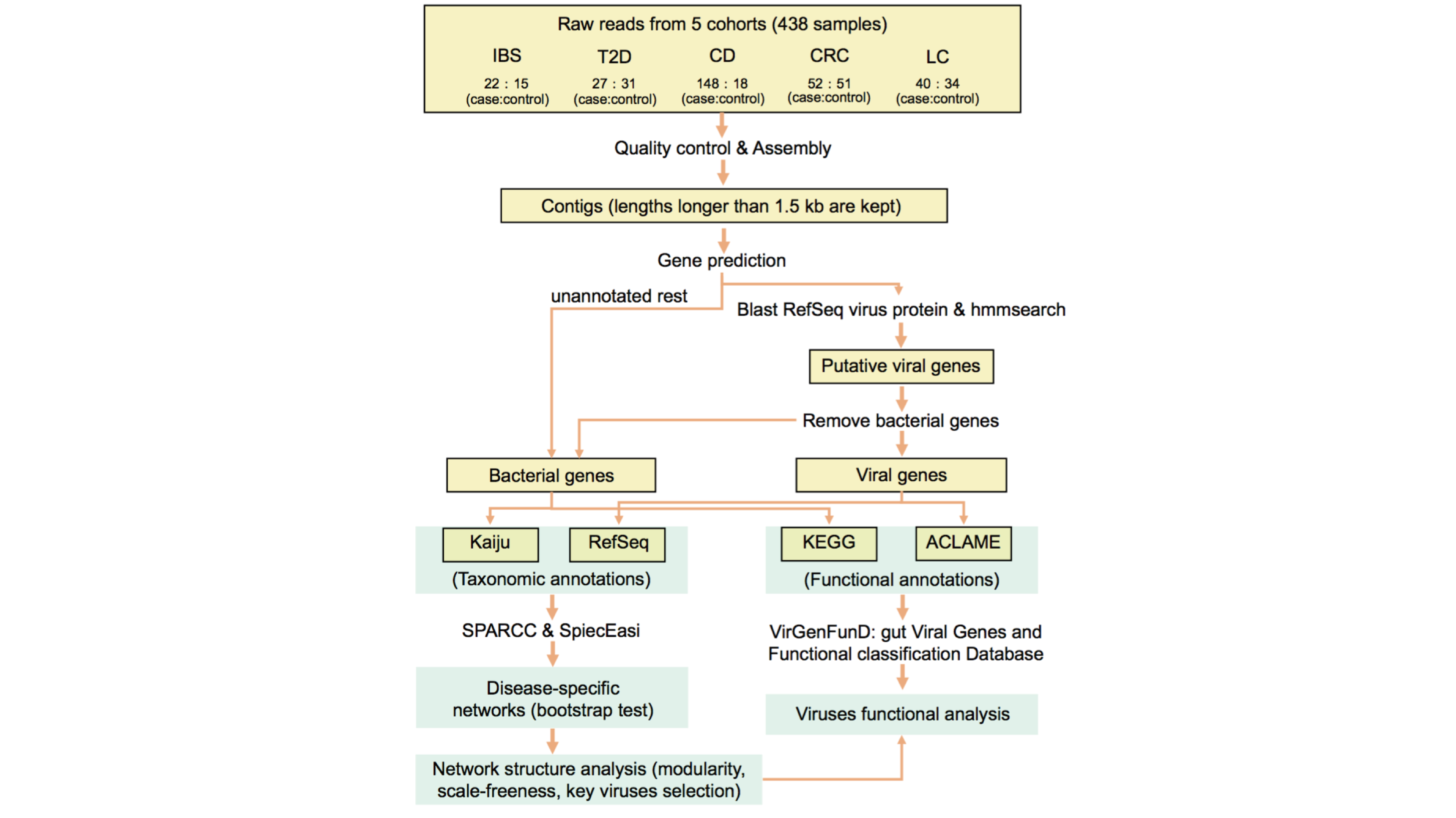
VirGenFunD (gut Viral Genes and Functional classification Database) is a database of disease-related viral genes with multi-level annotations. With well-designed pipeline for precise viral genes identification, the current version of VirGenFunD contains 3,351,765 viral gene sequences detected in metagenomics datasets of five diseases and the corresponding healthy controls: irritable bowel syndrome (IBS), type 2 diabetes (T2D), Crohn’s disease (CD), colorectal cancer (CRC) and liver cirrhosis (LC). We manually classified viral gene functions into 16 categories to facilitate systematic view of function of viruses in human gut. We provide the search function for the annotation of ACLAME protein family (all).
Importance and Applications
- To gain understanding of the function of viral protein families.
- To understand the viral gene functions in human gut.
- To serve as references of human gut and disease related gut viral gene catalog.
- To aid the identification of viral markers between diseases and health.
Construction of VirGenFunD and related analyses pipeline
VirGenFunD statistics
Comparison of number of annotations for viral sequences in VirGenFunD:
Publication
Mo Li, Chunhui Wang, Xiaoqing Jiang, Qian Guo, Congmin Xu, Zhongjie Xie, Peihong Wang, Jie Tan, Shufang Wu, Jinyuan Guo, Zhencheng Fang, Shiwei Zhu, Liping Duan, Huaiqiu Zhu. Characterization of human gut virome by analysis of multi-diseases gut metagenomes and disease-specific interactions.